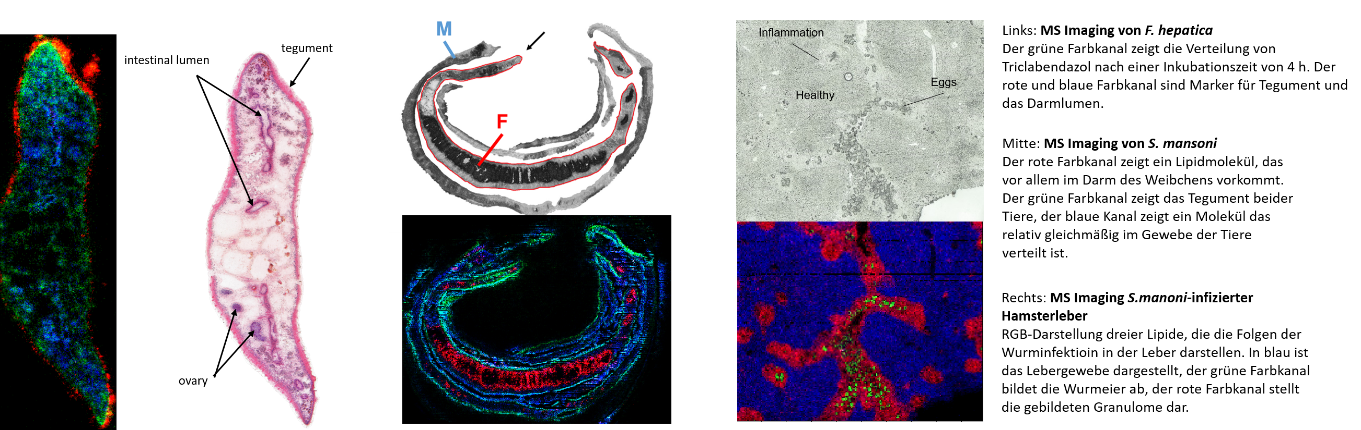
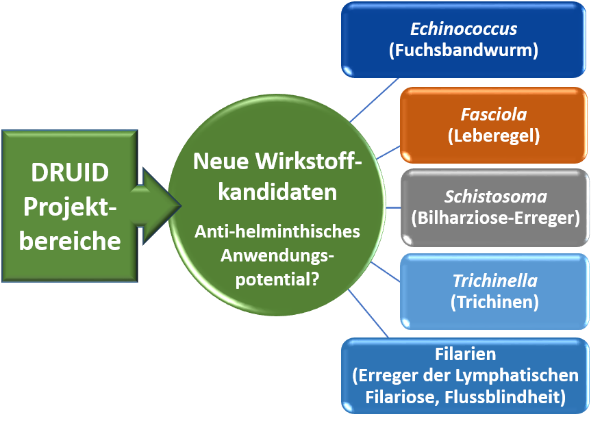
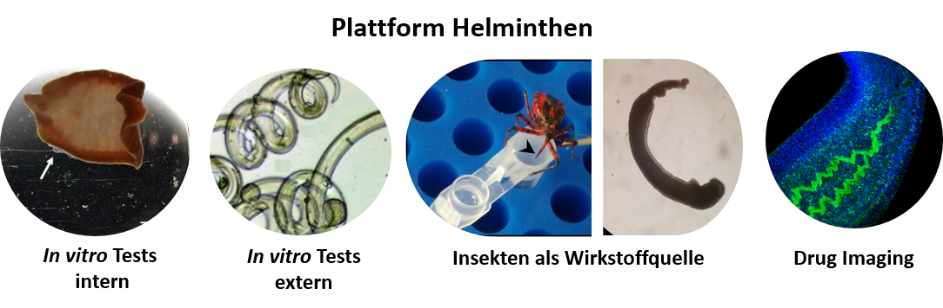
Process development/ Process control (PAT, cGMP), Production platform for proteins and virus-like particles
Project description:
For a translation of research results from the DRUID consortium into the clinic or industry, robust production processes, which represent more than just scaling up, are essential. These processes must follow the guidelines of good manufacturing practice (cGMP) and process analytical technology (PAT). The following work is planned: i) process intensification and expansion of the BEVS production platform as well as implementation of a continuous process control, ii) integration of online PAT technology, e.g., impedance spectroscopy for an automated determination of the optimal point in time for harvesting and automated harvesting, iii) expression (BEVS) and purification of S. mansoni kinases as well as study of the kinases and putative inhibitors, and iv) study of novel transfection reagents for transient protein production on a bioreactor scale.
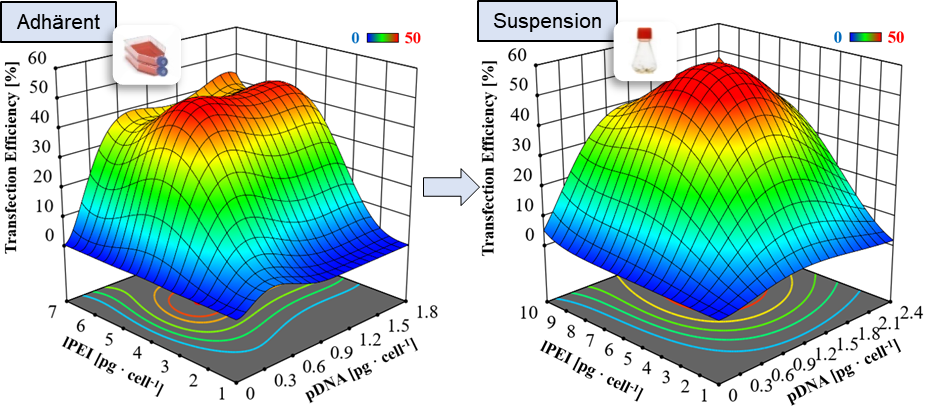
Optimization of the transient transfection process using statistical design of experiments. ©IBPT
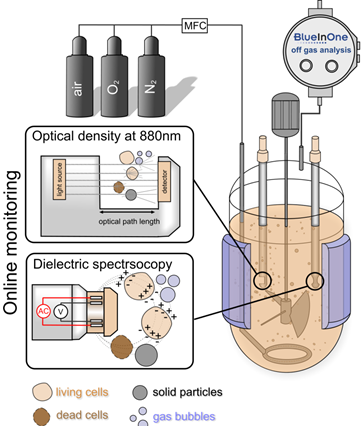
Production concept with integrated PAT technology. ©IBPT
Scientific goal:
The work on production process development and process control will be further adapted to the questions of the center, expanded, and offered to the entire consortium for use. It is planned to further intensify and automate the established BEVS (Baculovirus Expression System) platform. In addition, the second production platform – the transient production with HEK-293T cells – is also being further developed.
DRUID Collaboration partners:
B4 Schlitzer Lab, B5 Grevelding lab, B7 P Falcone Lab, C6 NWG Häberlein, E1 Platform Grevelding/ Häberlein
References E5: Biotechnol, DOI: 10.1016/j.ejbt.2021.08.002, 3. Lothert et al. (2020) Methods Mol Biol 2183:217-248, 4. Dekevic et al. (2022) J Biotechn 346 23-34, 5. Schwarz et al. (2021) Elec J Biotechnol, DOI: 10.1016/j.ejbt.2022.01.003; 6. Barekzai et al. (2020) New Adv Ferm Processes, DOI: 10.5772/intechopen.90029, 7. Eckhardt et al. (2021) Sep Sci Technol, 57 (6) 886-897