Project area D:
D1Inhibition of virus-activating host proteases
Eva Friebertshäuser, Torsten Steinmetzer
Prof. Dr. Eva Friebertshäuser
Institut für Virologie
Philipps-Universität Marburg
Hans-Meerwein-Str. 2
35043 Marburg
Tel.: +49 (0)6421-28 66019
Fax: +49 (0)6421-28 68962
E-Mail: friebertshaeuser(at)staff.uni-marburg(dot)de
Prof. Dr. Thorsten Steinmetzer
Institut für Pharmazeutische Chemie
Philipps-Universität Marburg
Marbacher Weg 10
35032 Marburg
Tel.: +49 (0)6421-28 25900
Fax: +49 (0)6421-28 25901
E-Mail: torsten.steinmetzer(at)staff.uni-marburg(dot)de
Project description:
Cleavage of viral envelope proteins by host proteases is essential for the infectivity of many human pathogenic viruses. Among others, the surface glycoproteins of highly pathogenic avian influenza viruses (e.g. H5N1), chikungunya virus or dengue, West Nile and Zika viruses are activated by furin-like serine proteases. The surface glycoproteins hemagglutinin of zoonotic H7N9 and seasonal influenza A viruses or the spike protein S of many coronaviruses (CoV) are cleaved by the membrane-bound trypsin-like serine protease TMPRSS2. Recently, we were able to show that SARS-CoV-2 S is activated by both furin and TMPRSS2. Therefore, these host proteases are promising targets for the development of novel broad-spectrum antiviral agents.
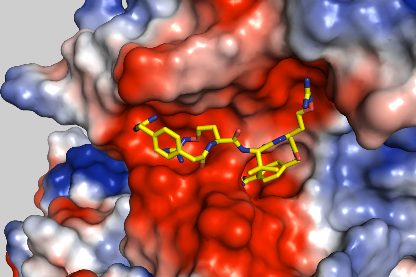
Crystal structure of furin in complex with inhibitor MI-1851

Crystal structure of TMPRSS2 superimposed with inhibitors MI-1904 (yellow) and MI-432 (orange).
Scientific goal:
Structure-based development of effective inhibitors targeting virus-activating host proteases; determination of their potency and selectivity by enzyme kinetic studies; structural characterization of their binding mode in complex with relevant host proteases; testing of their antiviral efficacy against significant human pathogenic viruses in cell cultures, tissue cultures and animal models.
DRUID Collaboration partners:
A1 Becker lab, A2 Grünweller lab, B6 Herker lab, C1 Hildt lab
References D1: 1. Böttcher et al. (2006) J Virol 80: 9896-8 3. Becker et al. (2012) J Biol Chem 287: 21992-03 4. Böttcher-Friebertshäuser et al. (2012) Vaccine 30: 7374-80 5. Ivanova et al. (2017) ChemMedChem 12: 1953-68 6. Lam van et al. (2019) ChemMedChem 14, 673-85 7. Bestle et al. (2020) LSA 3: e202000786 8. Bestle et al. (2021) J Virol 95: e0090621 9. Lam van et al. (2021) ACS Med Chem Lett 12: 426-32.
D2HEV, Identification of cellular targets for antiviral strategies-inhibition of virus release by modulation of cholesterol level
Kai-Henrik Peiffer, Stefan Zeuzem, Eberhard Hildt
Dr. Kai-Henrik Peiffer
Goethe-Universität Frankfurt am Main
Universitätsklinikum
Zentrum der Inneren Medizin
Medizinische Klinik I
Theodor-Stern-Kai 7
60596 Frankfurt am Main
E-Mail: kai-henrik.peiffer(at)kgu(dot)de
Prof. Dr. Eberhard Hildt
Bundesinstitut für Impfstoffe
und biomedizinische Arzneimittel
Paul-Ehrlich-Institut
Paul-Ehrlich-Straße 51-59
63225 Langen
Tel.: +49 (0)6103-77 2140
Fax: +49 (0)6103-77 1234
E-Mail: Eberhard.Hildt(at)pei(dot)de
Prof. Dr. Stefan Zeuzem
Department of Medicine
Goethe University Hospital
Theodor-Stern-Kai 7
60590 Frankfurt am Main
Tel.: +49 (0)69-6301 4544
E-Mail: zeuzem(at)em.uni-frankfurt(dot)de
Project description:
HEV infects more than 20 Mio people per year and concerns industrial nations as well as developing countries. At present there is no specific therapy available.As non-enveloped virus HEV release depends on endodomal processes which represent a promising target for antivirals. In the first funding period the lysosmal degradation of HEV in the endosomal system was identified as a central anchor point for antivirals. In addition to innate immunity cholesterol homeostasis is a a central factor affecting HEV reelase. Based on this the underlying mechanisms are analysed in detalil to identifiy further targets which can be addressed by drug repurposing. Animal models will help to broaden the insight in immunological processes controlling HEV life cycle.
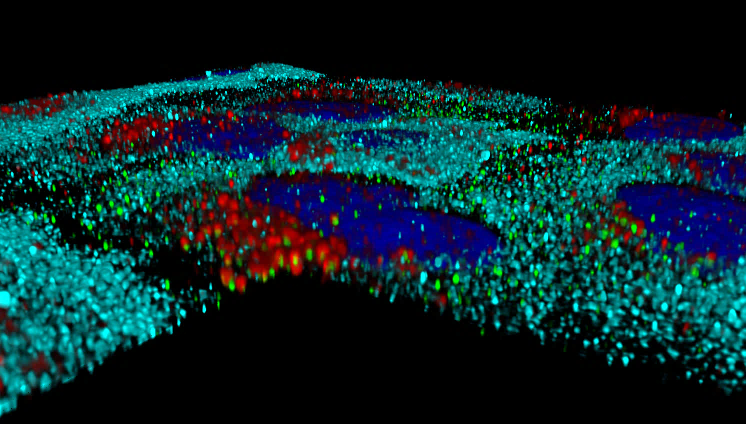
3D reconstruction of subcellular distribution of HEV (green) , GBP-1 (cyan) and lysosomes (red) in interferon -treated cells.
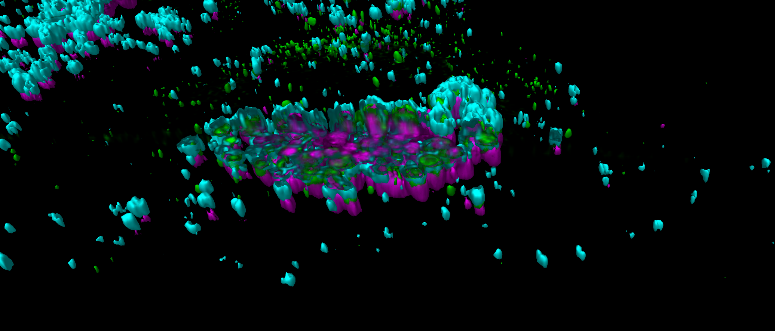
CLSM-based 3D reconstruction of sub-cellular distribution of HEV (green) and lysosomes (cyan) after induction of and cholesterol accumulation (magenta).
Scientific goal:
Inhibtion of the endosomal life cycle of HEV by modulation of cholesterol-dependent regulated target structurs via application of identified bioactive compounds (drug repurposing).
DRUID Collaboration partners:
A2 Grünweller lab, A4 Heine/Reuter lab, B1 Diederich/Kolb lab, B6P Herker lab, C5 Glebe/Geyer lab, D1 Steinmetzer lab
References D2: [1] Glitscher et al. (2018) Viruses 10(6):301; [2] Müller et al. (2020) Antiviral Res 174:104706; [3] Basic et al. (2019) Antiviral Res 172:104644; [4] Glitscher et al. (2021) J Virol doi: 10.1128/JVI.01564-20; [5] Glitscher et al. (2021) Cell Mol Gastroenterol Hepatol, doi:10.1016 /j.jcmgh.2021.02.002; [6] Himmelsbach et al. (2018) Emerg Microbes Infect 7(1):196. [7] Glitscher et al. (2021) Cell Microbiol. 16:e13379
D3Characterisation of apoptotic parasites to identify new target molecules for the treatment of Leishmaniasis
Ger van Zandbergen
Prof. Dr. Ger van Zandbergen
Abteilung Immunologie
Paul-Ehrlich-Institut
Paul-Ehrlich-Str. 51-59
63225 Langen
Tel.: +49 (0)6103-77 2005
E-Mail: Ger.Zandbergen(at)pei(dot)de
Project description:
Leishmaniasis is a neglected tropical disease caused by the parasite Leishmania spp, which is endemic in almost 100 countries. We could demonstrate that apoptotic Leishmania major (L. major) promastigotes are both responsible for the infectivity and the survival of parasites in host cells. Apoptotic parasites induce an anti-inflammatory response in human macrophages that does not lead to an effective T-cell response against Leishmania. Even if L. major parasites show all typical characteristics of apoptosis, typical eukaryotic apoptosis regulating proteins are not present in Leishmania.
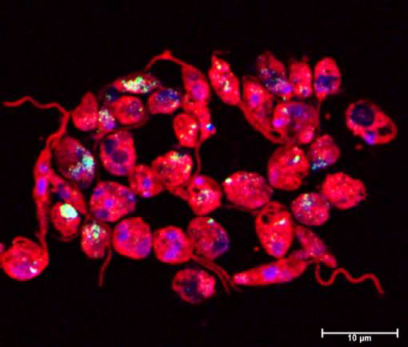
Staining of fragmented DNA (TUNEL staining) in L. major Cas9/T7 in which apoptosis was induced by Miltefosin (TUNEL: green; DAPI: blue; anti-Lm-serum: red. © Ger van Zandbergen)
Scientific goal:
For a novel vaccination approach and to kill parasites we aim to identify apoptosis regulating proteins in Leishmania as potential drug targets as well as attenuated Leishmania strains without anti-inflammatory properties.
DRUID Collaboration partners:
B1 Kolb, B3 Rahlfs/Kolb, E3 Rahlfs/Przyborski
References D3: [1] Arens et al. (2018) Front Immunol 31(9):1772. [2] Crauwels et al. (2019) Front Immunol 22(10):2697. Further publications within DRUID: [3] Turoňová et al. (2020) Science 370(6513):203-208.
D4Blockage of glutaminolysis and glycolysis for Cryptosporidium parvum inhibition and One Health study on cryptosporidiosis in Cameroon
Carlos Hermosilla, Anja Taubert, Sybille Mazurek
Prof. Dr. Sybille Mazurek
Institut für Veterinär-Physiologie und -Biochemie
Fachbereich Veterinärmedizin
Justus-Liebig-Universität Gießen
Frankfurter Str. 100
35392 Gießen
Tel.: + 49 (0)641-99 38182
E-Mail Sybille.Mazurek(at)vetmed.uni-giessen(dot)de
Prof. Dr. Carlos Hermosilla
Institut für Parasitologie/Institut für Veterinär-Physiologie und -Biochemie
Justus-Liebig-Universität Gießen
Schubertstr. 81/Frankfurter Str. 100
Tel.: +49 (o)641-99 38461/99 38182
E-Mail: Carlos.R.Hermosilla(at)vetmed.uni-giessen(dot)de
Prof. Dr. Anja Taubert
BFS, Institut für Parasitologie
Justus-Liebig-Universität Gießen
Schubertstraße 81
35392 Gießen
Tel.: +49 (0)641-99 38460
Fax: +49 (0)641-99 38469
E-Mail: Anja.Taubert(at)vetmed.uni-giessen(dot)de
Project description:
Cryptosporidium spp. are intestinal parasites which cause severe diarrhoea in humans, especially in HIV patients and young children. Especially in developing countries these protozoan infections induce high morbiditiy and mortality. So far, epidemiological factors contributing to cryptosporidiosis in underdeveloped countries – such as Cameroon – have insufficiently been studied. Currently available therapeutics show insufficient efficacies in the above mentioned risk groups. Cryptosporidium spp. are obligate intracellular protozoa which own minimal metabolic capacities. Consequently, to sustain their intracellular proliferation, these parasites have to modulate the host cellular metabolism. By characterizing metabolic signatures of C. parvum-infected host cells, we recently identified host cell metabolic reactions and pathways that are essential for parasite proliferation.
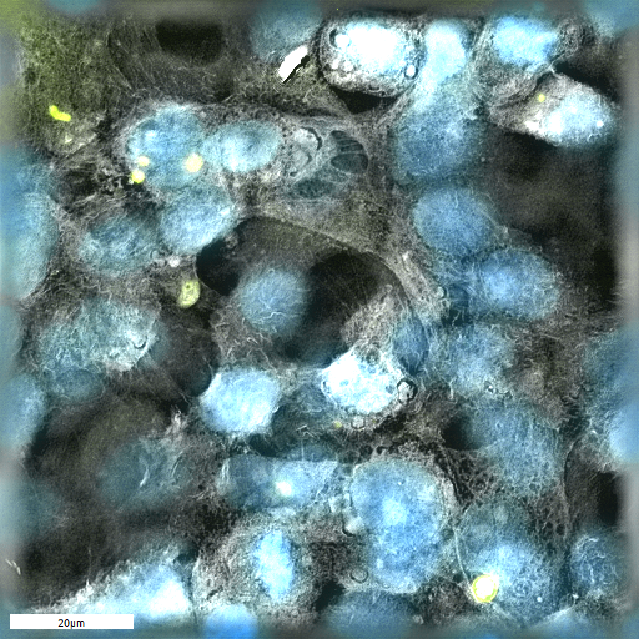
Cryptosporidium parvum- (yellow) infected host cells (nuclei: blue), tomographic microscopie © Juan Vélez
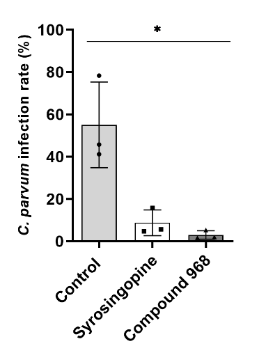
Inhibition of Cryptosporidium parvum via exemplary metabolic blockers (Vélez et al. 2021c)
Scientific goal:
This projects targets specific metabolic pathways of host cells (mainly glycolysis and glutaminolysis) by using new metabolic inhibitors or combinatory treatments. Moreover, a One Health study on several epidemiological factors of cryptosporidiosis in Cameroon is conducted.
DRUID Collaboration partners:
E4 Spengler lab
References D4: 1. *Vélez et al. (2021) Pathogens 11(1):49. 2. * Vélez et al. (2021) Biology 10(10):963 3. ** Vélez et al. (2021) Biology 10(1):60








