Project area E:
E1Platform Drug Testing Against Helminths
Simone Häberlein, Christoph Grevelding
PD Dr. Simone Häberlein
Justus-Liebig-Universität Gießen
BFS, Institut für Parasitologie
Schubertstraße 81
35392 Gießen
Tel.: +49 (0)641-99 38476
Fax: +49 (0)641-99 38469
E-Mail: Simone.Haeberlein(at)vetmed.uni-giessen(dot)de
Prof. Dr. Christoph Grevelding
BFS, Institut für Parasitologie
Justus-Liebig-Universität Gießen
Schubertstraße 81
35392 Gießen
Tel.: +49 (0)641-99 38466
Fax: +49 (0)641-99 38469
E-Mail: Christoph.Grevelding(at)vetmed.uni-giessen(dot)de
Project description:
Targets of antiparasitic compounds are often conserved, which opens the possibility to achieve therapy of different parasitic infections with the same active compound. In platform project E1, we focus on parasitic worms (helminths), for which the discovery of new active compounds is particularly important because of resistances or suboptimal therapeutic successes of existing drugs. To this end, we implement in vitro screenings of compounds originating from DRUID projects to test their efficacy against the blood fluke Schistosoma mansoni (causing schistosomiasis) and the liver fluke Fasciola hepatica (causing fascioliasis). The platform project cooperates with several national and international partners, including colleagues in endemic countries in Asia and Africa, who open us test possibilities against numerous other helminths species.
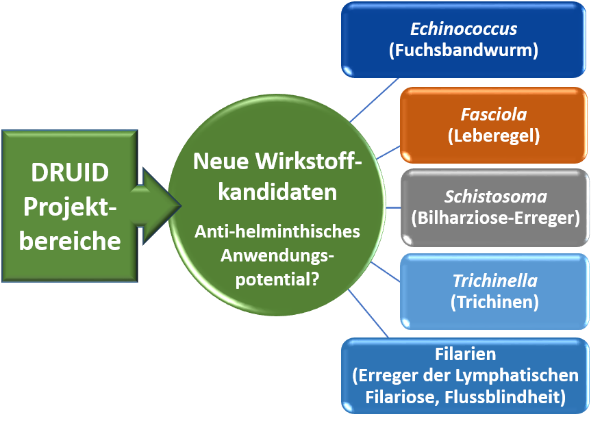
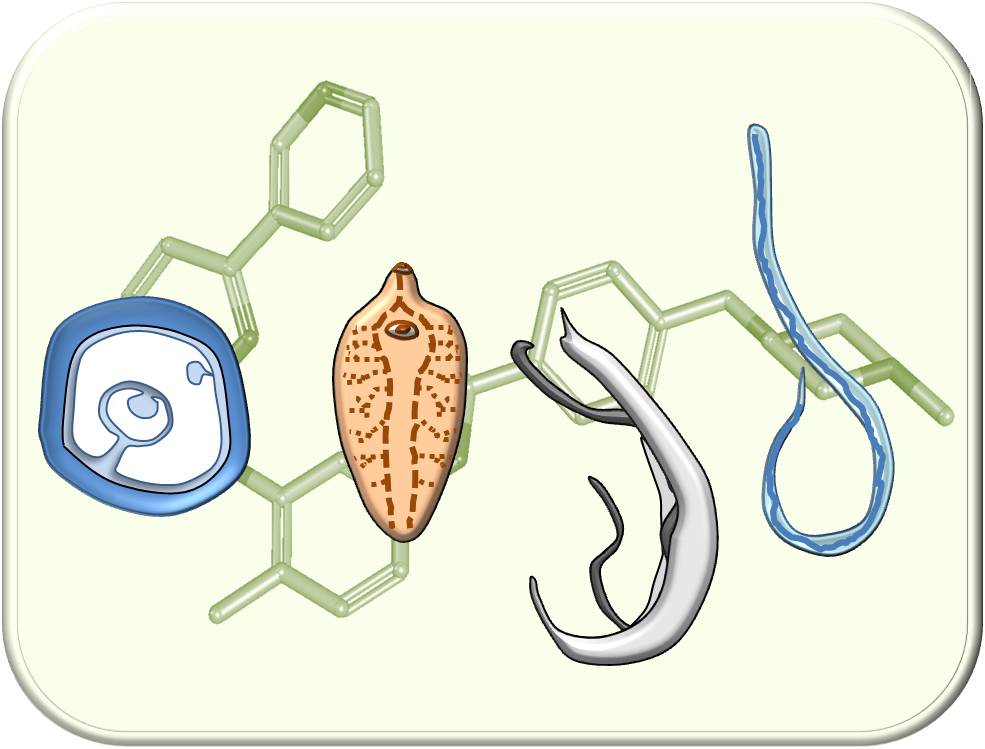
Strategy for identifying active compounds with broad antiparasitic efficacy against globally important helminths species. ©Simone Häberlein
We also investigate the mechanism of action for selected test compounds by applying various in vitro culture-based and imaging methods.
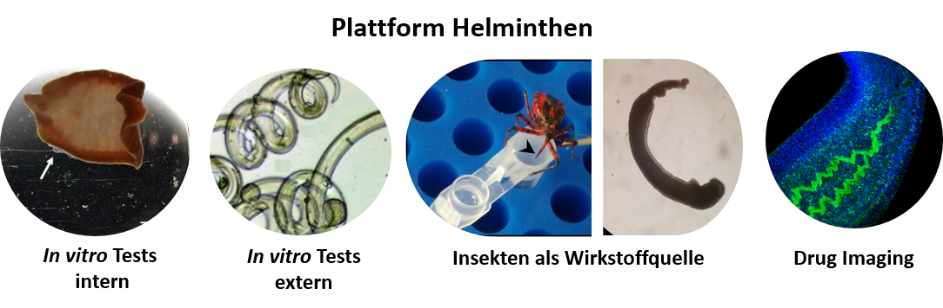
Test spectrum provided by the platform project to identify and characterize anthelminthic compounds. ©Simone Häberlein, Miray Tonk-Rügen
Scientific goal:
Aim of the project is the identification of substances with a broad anti-parasitic efficacy and the discovery of conserved modes of action.
DRUID Collaboration partners:
A2 Grünweller lab, A6 Douglas lab, A7 Przyborski lab, B4 Schlitzer lab, B5 Grevelding lab, B7 Falcone lab, E4 Spengler lab
References E1: 1. Peter-Ventura et al. (2019) ChemMedChem 14(21):1856-1862. 2. Houhou et al. (2019) Sci Rep 9:15867. 3. Li et al. (2019) Parasitol Res 118(3):881-890. 4. Morawietz et al. (2020) Front Vet Sci 7:611270. 5. Kellershohn et al. (2019) PLOS Negl Trop Dis 13:e0007240. 6. Tonk et al. (2020) Antibiotics 9:664. 7. Mokosch et al. Anal Bioanal Chem 413(10): 2755-2766. 8. Mughal et al. (2021a) Int J Parasitol 51(7):571-585. 9. Mughal et al. (2021b) Int J Parasitol S0020-7519(21)00312-X. 10. Gallinger et al. (2022) Pharmaceuticals 15(2): 119. 11. Morawietz et al. (2022) Parasitol Res (online ahead of print) doi: 10.1007/s00436-021-07388-1
E3Protein production, assay development, crystallisation and interaction analysis
Jude Przyborski, Stefan Rahlfs
Prof. Dr. Jude Przyborski
Interdisziplinäres Forschungszentrum (iFZ)
Justus-Liebig-Universität Gießen
Heinrich-Buff-Ring 26-32
35392 Gießen
Tel.: +49 (0)641-99 39114
E-Mail: jude.przyborski(at)ernaehrung.uni-giessen(dot)de
Dr. Stefan Rahlfs
Biochemie und Molekularbiologie
Justus-Liebig-Universität Gießen
Heinrich-Buff-Ring 26-32
35390 Gießen
Tel.: +49 (0)641-99 39117
E-Mail: stefan.rahlfs(at)ernaehrung.uni-giessen(dot)de
Project description:
Within the framework of this platform project, we offer methods for protein production, for the development of high-throughput assays, for crystallisation and structural analysis as well as for interaction analyses. A variety of optimisation options for cloning/expression and purification of native proteins (affinity chromatography, gel filtration, ion exchange) are available. Once recombinant protein has been obtained, provide support in the development of assay systems up to high throughput formats. Furthermore, the platform proves access to the crystallisation platform at the iFZ. A pipetting and a crystallisation robot as well as starter kits and various additive screens for optimisation are available for crystallisation screens on the nl scale. The system is currently being updated with funding from the latest DRUID funding round. Dr Fritz-Wolf acts as crystallographer and contact person and can carry out the first tests on the crystals. The X-ray sources of the Max Planck Institute for Medical Research (Heidelberg) and the AG Klebe/Heine [A5] (Marburg) are available for this purpose.
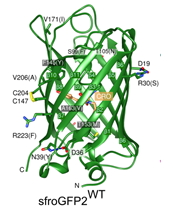
Crystal structure of sfroGFP (Heimsch et al. 2022)
DRUID Collaboration partners:
Protein-Expression: A1, A3, A6NWG, A7, B2, B3, B4, C3
Crystallisation: B2, B3, C3
Assay-Devlopment: A7, B3
References E3: 1. Heimsch et al. (2022) Antioxid & Redox Signal doi: 10.1089/ars.2021.0234 2. Harnischfeger et al. (2020) Electronic Journal of Biotechnology 3. Fritz-Wolf et al. (2011) Nat Commun 2:383 4. Koncarevic et al. (2009) Proc Natl Acad Sci USA 106:13323-8
E4High resolution mass spectrometry imaging
Bernhard Spengler
Prof. Dr. Bernhard Spengler
Institut für Anorganische und Analytische Chemie
Justus-Liebig-Universität Gießen
Heinrich-Buff-Ring 17
35392 Gießen
Tel.: +49 (0)641-99 34801
E-Mail: bernhard.spengler(at)ac.jlug(dot)de
Project description:
High resolution mass spectrometry imaging provides numerous opportunities to aid in the characterization of pathogens, host-pathogen interactions and the analysis of drug molecules. The sample is analyzed with a laser in a rasterized fashion, the step size is only a few micrometers. The sample material is ablated and ionized, allowing for the parallel detection of hundreds of chemical compounds as well as the creation of images that show the distribution of the analytes throughout the sample. Identification of the molecules is carried out using exact mass determination and is supported by fragmentation experiments. Continued improvements in instrumentation allow for analysis and graphical representation of smaller structures while additional sample preparation steps like in-situ derivatization provide the opportunity to detect analytes at very low concentrations.
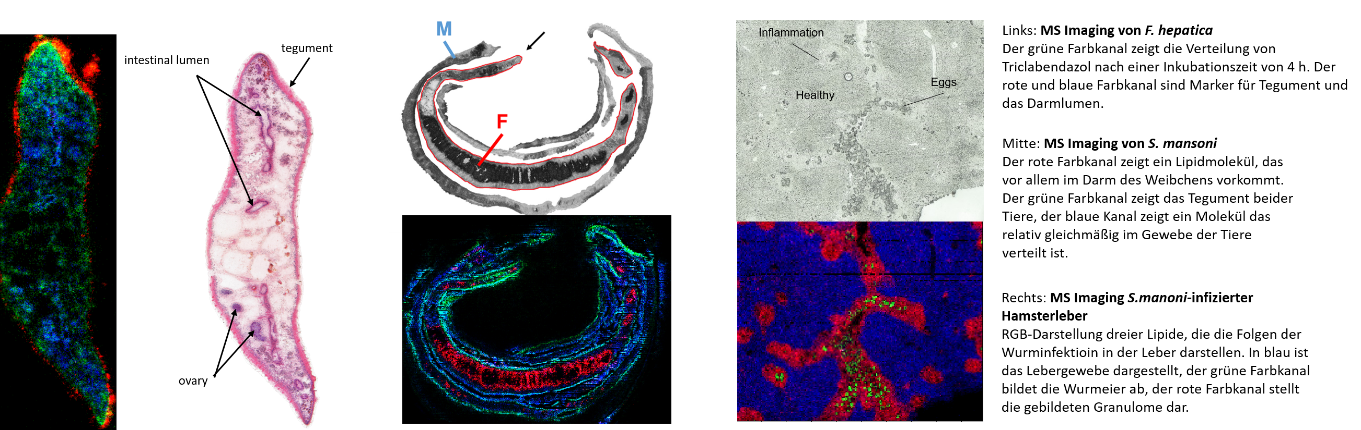
Scientific goal:
The goal of the project is the improvement and application of the available methods for the demands of the DRUID center. Especially the distribution of drug compounds, the lipidomic and metabolomic characterization of pathogens and the interaction of pathogens and hosts are important research areas.
DRUID Collaboration partners:
A2 Grünweller lab, B4 Grevelding lab, B5 Schlitzer lab, D4 Taubert lab, E1 Häberlein lab, W3 Herker lab
References E4: [1] Kadesch et al. (2020) PLoS Negl. Trop. Dis. 14(5): e0008145; [2] Morawietz et al. (2020), Front. Vet. Sci. 7, 611270; [3] Mokosch et al. (2021) Anal Bioanal Chem 413, 2755–2766; [4] Müller et al. (2021) J. Am. Soc. Mass Spectrom.,3,32(2),465-472; [5] Spengler et al. (2015) Anal Chem 87 (1) 64-82.
E5Process development/ Process control (PAT, cGMP), Production platform for proteins and virus-like particles
Peter Czermak, Denise Salzig
Prof. Dr. Ing. Peter Czermak
Institut für Bioprozesstechnik
Technische Hochschule Mittelhessen
Wiesenstraße 14
35390 Gießen
Tel.: +49 (0)641-309 2551
E-Mail: peter.czermak(at)lse.thm(dot)de
Prof. Dr. Ing. Denise Salzig
Institut für Bioverfahrenstechnik
und Pharmazeutische Technologie
Technische Hochschule Mittelhessen
Gutfleischstr. 3-5
Tel.: +49 (0)641-309 2630
Fax: +49 (0)641-309 2553
E-Mail: denise.salzig(at)lse.thm(dot)de
Project description:
For a translation of research results from the DRUID consortium into the clinic or industry, robust production processes, which represent more than just scaling up, are essential. These processes must follow the guidelines of good manufacturing practice (cGMP) and process analytical technology (PAT). The following work is planned: i) process intensification and expansion of the BEVS production platform as well as implementation of a continuous process control, ii) integration of online PAT technology, e.g., impedance spectroscopy for an automated determination of the optimal point in time for harvesting and automated harvesting, iii) expression (BEVS) and purification of S. mansoni kinases as well as study of the kinases and putative inhibitors, and iv) study of novel transfection reagents for transient protein production on a bioreactor scale.
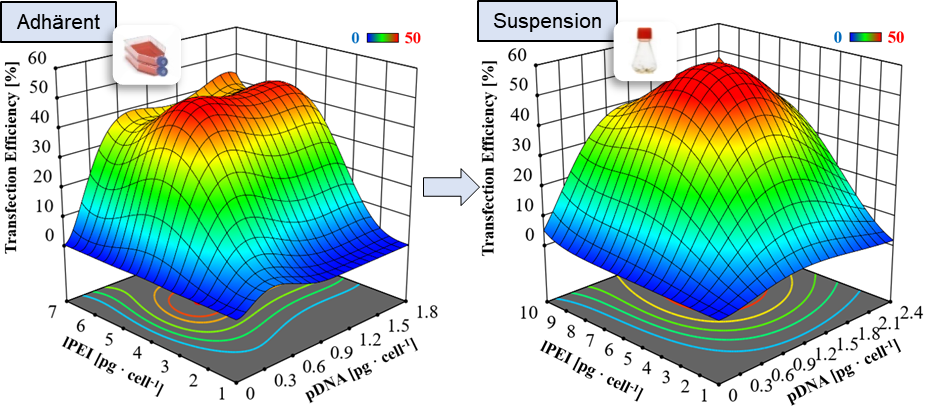
Optimization of the transient transfection process using statistical design of experiments. ©IBPT
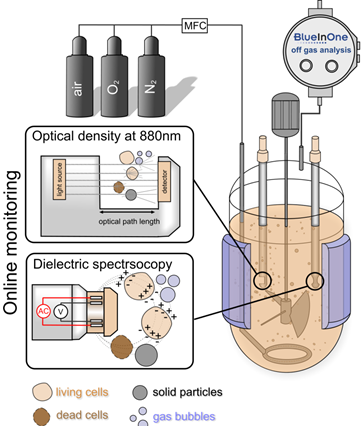
Production concept with integrated PAT technology. ©IBPT
Scientific goal:
The work on production process development and process control will be further adapted to the questions of the center, expanded, and offered to the entire consortium for use. It is planned to further intensify and automate the established BEVS (Baculovirus Expression System) platform. In addition, the second production platform – the transient production with HEK-293T cells – is also being further developed.
DRUID Collaboration partners:
B4 Schlitzer Lab, B5 Grevelding lab, B7 P Falcone Lab, C6 NWG Häberlein, E1 Platform Grevelding/ Häberlein
References E5: Biotechnol, DOI: 10.1016/j.ejbt.2021.08.002, 3. Lothert et al. (2020) Methods Mol Biol 2183:217-248, 4. Dekevic et al. (2022) J Biotechn 346 23-34, 5. Schwarz et al. (2021) Elec J Biotechnol, DOI: 10.1016/j.ejbt.2022.01.003; 6. Barekzai et al. (2020) New Adv Ferm Processes, DOI: 10.5772/intechopen.90029, 7. Eckhardt et al. (2021) Sep Sci Technol, 57 (6) 886-897
E6Pharmaceutical development, translational medicine and drug repurposing
Susanne Schiffmann
Dr. Susanne Schiffmann
Fraunhofer Institut für Translationale Medizin und Pharmakologie ITMP
Theodor-Stern-Kai 7
60596 Frankfurt am Main
Tel.: +49 (0)69-8700 25060
E-Mail: susanne.schiffmann(at)itmp.fraunhofer(dot)de
Project description:
The SARS-CoV2 pandemic clearly demonstrates that there is an unmet medical need for anti-pathogenic drugs. In order to identify new treatment options, promising substances from other working groups will be characterized preclinical. This includes the generation of a safety profile and/or bioavailability testing of the drug candidates. Since supportive therapy with immune-modulating drugs is a proven therapeutic approach, an interaction of the drug candidates with the immune system will be tested.
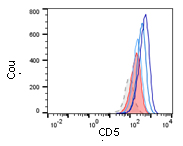
Surface marker expression of treated M2 macrophages ©Marina Henke
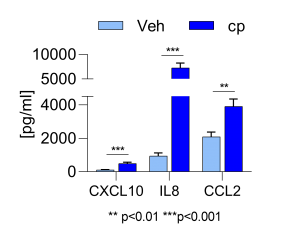
Cytokine expression of treated M2 macrophages. ©Leonard Blum
Scientific goal:
As part of the project, a safety and/or immunomodulatory profile will be created for small molecules/peptides that have a potential antiviral, antiparasitic or antibacterial property. Furthermore, human proteins (e.g. TMPRSS2) which represent new targets for combating pathogens will be investigated.
DRUID Collaboration partners:
A1 Becker lab, A2 Grünweller lab, A3 Weber lab, B1 Diederich/Kolb lab, B3 Rahlfs/Kolb/van Zandbergen, B5 Schlitzer lab, D1 Friebertshäuser lab
References E6: 1. Blum et al. (2021) J Mol Med 99(2):261-72; 2. Blum et al. (2020) Sci Rep 10(1):7534; 3. Blum et al. (2020) J Cell Mol Med 24(12):6988-99.
E7 PNeglected infectious diseases with focus on imaging
Jacomina Krijnse Locker
Prof. Dr. Jacomina Krijnse Locker
Elektronenmikroskopie von Pathogenen
Paul-Ehrlich-Institut
Paul-Ehrlich-Straße 51-59
63225 Langen
Tel.: +49 (0)6103-77 2011
E-Mail: Jacomina.KrijnseLocker(at)pei(dot)de
Project description:
We apply imaging techniques to understand how pathogens interact with cells to cause disease, focusing on electron microscopy (EM) and correlated light- and electron microscopy (CLEM). To generate robust protocols that can easily be adapted, we use our model virus, the large DNA-virus vaccinia (VACV).
The SARS CoV2 pandemic illustrates how viruses have a world-wide socio-economical impact with dramatic consequences for the neglected tropical diseases (NTDs). There is an urgent need to contain the SARS CoV2 pandemic and refocus resources on NTDs. In a collaborative effort we showed that the spike-surface protein (S) of SARS COV2, that mediates infection, shows unexpected flexibility. The latter could facilitate receptor binding and cell entry of the virus, which remains to demonstrated.
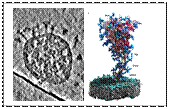
Cryo-electron tomography and molecular dynamic simulation of SARS-CoV-2 spike shows unexpected flexibility of the stalk.
Upon budding at intracellular membranes coronaviruses leave the cell in a poorly explored way. Vesicular trafficking as well as regulated lysosomal exocytosis have been proposed.
Scientific goal:
The project aims to interfere with the flexibility of S and test for the outcome of infection using antibodies, drugs and genetics. Virus release from infected cells will be studied by a CLEM approach, combining live cell imaging with EM. The expertise gained from our VACV- and SARS CoV2 host system is then applied to answer questions related to virus-host systems used in other teams of the DRUID-consortium.
DRUID Collaboration partners:
A3 Weber lab, B2 Ziebuhr lab, B6 P Herker lab, C1 Hildt lab, D3 van Zandbergen lab, E5 Czermak lab
References E7 P: Turoňová, B., et al. (2020). Science (80-. ). 370, 203–208. Blanco-Rodriguez, G., et al. (2020). J. Virol. 94 e00135-20. Quemin, E.R., et al. (2018). J. Mol. Biol.430, 1714-1724. Chlanda, P., and Krijnse Locker, J. (2017). Biochem. J. 474, 1041–1053. Sartori-Rupp, A., et al. (2019). Nat. Commun. 10, 342.









