Project area B:
B1Dengue and Zika virus, design of inhibitors of NS3/NS2B serine protease.
Wibke Diederich, Peter Kolb
Prof. Dr. Wibke Diederich
Institut für Pharmazeutische Chemie
Zentrum für Tumor- und Immunbiologie
Philipps-Universität Marburg
Hans-Meerwein-Straße 3
35043 Marburg
Tel.: +49 (0)6421-28 25810
Fax: +49 (0)6421-28 26254
E-Mail: wibke.diederich(at)staff.uni-marburg(dot)de
Prof. Dr. Peter Kolb
Institut für Pharmazeutische Chemie
Philipps-Universität Marburg
Marbacher Weg 8
35032 Marburg
Tel.: +49 (0)6421-28 25908
Fax: +49 (0)6421-28 26652
E-Mail: peter.kolb(at)uni-marburg(dot)de
Project description:
Infections with the dengue virus have reached a new high in recent years, with around 50-100 million new infections per year. In the vast majority of cases, the infection progresses with mild, flu-like symptoms, but a small percentage of those affected, often children, develop hemorrhagic fever, which is fatal if severe. Although a vaccine is now available, since this is only approved for a very limited group of people, the development of agents that efficiently suppress the multiplication of the virus is essential. Our work focuses on the virus’s own serine protease NS3/NS2B, which cleaves the viral precursor protein into functional proteins and is essential for the maturation of the virus.
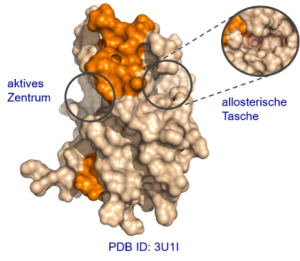
Abb. B1. Kristallstruktur der NS3/NS2B Protease DENV 3.1
Scientific goal:
The aim of the project is to further develop allosteric inhibitors of the viral serine protease NS3/NS2B, not only with respect to their affinity, but also with respect to their pharmacokinetic properties and toxicity (hit-to-lead development) using a combined approach of computer-aided design, synthesis, biological assays and crystal structure analysis. In addition, inhibitors of the related Zika protease will be developed based on the knowledge gained.
DRUID Collaboration partners:
A1 Becker, A2 Grünweller, B3 Rahlfs/Kolb/van Zandbergen, XY Herker, E6 Schiffmann/Laux
References B1: [1] Noble et al. (2012) J Virol 86(1):438-446; *[2] Chevillard et al. (2015), J Chem Inf Model 55(9):1824-1835; *[3] Chevillard et al. (2018) J Med Chem 61(3):1118-1129
B2Protein de-ADP-ribosylation und NMPylation activities as potential therapeutic targets against coronaviruses
John Ziebuhr
Prof. Dr. John Ziebuhr
Institut für Medizinische Virologie
Biomedizinisches Forschungszentrum
Seltersberg (BFS)
Justus-Liebig-Universität Gießen
Schubertstraße 81
35392 Gießen
Tel.: +49 (0)641-99 41200
Fax: +49 (0)641-99 41209
E-Mail: John.Ziebuhr(at)viro.med.uni-giessen(dot)de
Project description:
Coronaviruses are important human and animal pathogens. They are mainly associated with respiratory and intestinal infections and have significant zoonotic potential, resulting in several outbreaks of severe respiratory infections in humans over the past 2 decades including the SARS-CoV-2 pandemic starting in 2019. Therapeutic options to treat severe forms of COVID-19 and other coronavirus infections are very limited, indicating a high priority for the development of novel antiviral drugs. To address this need, project B2 focuses on two coronaviral proteins that are conserved among all coronaviruses: the coronavirus macrodomain (macD) in nonstructural protein 3 (nsp3) and the recently discovered nucleotidyltransferase (NiRAN) which is linked to the viral RNA-dependent RNA polymerase domain in nsp12 and was recently shown to be essential for coronavirus replication.
Coronavirus replicase polyprotein 1ab. Proteolytic processing by two or three viral proteases (PL, 3CL) results in the release of up to 16 nonstructural proteins (nsp) with numerous enzymatic and other functions.
Scientific goal:
The project aims to comprehensively characterize the biochemical properties of two coronavirus proteins/enzymes (macD, NiRAN) and their functions in the viral replication cycle using appropriate cell culture systems. Based on the conserved substrate specificity of the NiRAN domain, HTS assays will be developed and used to identify potential inhibitors.
DRUID Collaboration partners:
A2 Grünweller, A3 Weber, C5 Kraiczy, E3 Rahlfs/Przyborski
References B2: 1. *Slanina et al. (2021) Proc Natl Acad Sci USA 118: e2022310118 2. *Krichel et al. (2021) Sci. Adv. 7: eabf1004 3. *Müller et al. (2021) Antiviral Res 175: 1004706 4. *Snijder et al. (2016) Adv Virus Res 96: 59-126 5. *Putics et al. (2005) J Virol 79: 12721-31.
B3NAD(P)H-dependent metabolic pathways as targets for new anti-infective agents
Stefan Rahlfs, Ger van Zandbergen, Peter Kolb
Dr. Stefan Rahlfs
Biochemie und Molekularbiologie
Justus-Liebig-Universität Gießen
Heinrich-Buff-Ring 26-32
35390 Gießen
Tel.: +49 (0)641-99 39117
E-Mail: stefan.rahlfs(at)ernaehrung.uni-giessen(dot)de
Prof. Dr. Ger van Zandbergen
Abteilung Immunologie
Paul-Ehrlich-Institut
Paul-Ehrlich-Str. 51-59
63225 Langen
Tel.: +49 (0)6103-77 2005
E-Mail: Ger.Zandbergen(at)pei(dot)de
Prof. Dr. Peter Kolb
Institut für Pharmazeutische Chemie
Philipps-Universität Marburg
Marbacher Weg 8
35032 Marburg
Tel.: +49 (0)6421-28 25908
Fax: +49 (0)6421-28 26652
E-Mail: peter.kolb(at)uni-marburg(dot)de
Project description:
Cellular redox balance plays an essential role in pathogenic microorganisms. Enzymes of the NAD(P)H-dependent glutathione and thioredoxin system1,2, as well as the glucose 6-phosphate dehydrogenase (G6PD) and 6-phosphogluconate dehydrogenase (6PGD), which significantly contribute to the NADPH and ribose 5-phosphate pool via the pentose phosphate pathway3, are centrally involved. G6PD of the malaria parasite Plasmodium falciparum and P. vivax (GluPho), present as a bifunctional enzyme, differs functionally and structurally from the human host enzyme4 and is essential for malaria parasites5. Together with the Sanford-Burnham Institute/UCSD, La Jolla, we have established a high-throughput compatible assay for PfGluPho and have screened about 400,000 compounds (i.a. NIH MLSMR Collection)6. Following structure-activity relationship studies and lead optimization, we identified the highly selective PfGluPho inhibitor SBI-750, which is active in the lower nanomolar range7. The concept could already be transferred to Leishmania; the 3D crystal structure of Leishmania donovani G6PDs and PGDs could be solved and based on this a first in silico screening of small molecules could be realized. In addition, a high-throughput compatible assay for the recombinant G6PDs and 6PGDs from Leishmania was established to screen for inhibitors at the Novartis FAST lab in Cambridge (USA).
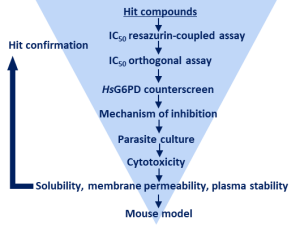
Workflow-Diagramm zur Inhibitor Identifizierung gegen G6PDs/PGDs
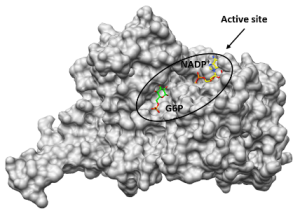
3D-Kristallstruktur LdG6PD ©Isabell Berneburg
Scientific goal:
The aim of this project is to functionally and structurally characterize the enzymes G6PD and 6PGD from Leishmania and Plasmodium as targets for drug development, and to identify and further develop inhibitors against these enzymes (in silico and HTS). The concept will also be transferred to other pathogens within the DRUID consortium, such as Schistosoma.
DRUID Collaboration partners:
B1 Diederich/Kolb; B4 Grevelding; D3 van Zandbergen; E1 Grevelding/Häberlein; E3 Rahlfs/Przyborski; E4 Spengler; E6 Schiffmann/Laux
References B3: 1. Fritz-Wolf et al. (2011) Nature Comm. 2:383* 2. Koncarevic et al. (2009) PNAS 106: 13323-8* 3. Bozdech and Ginsburg (2005) Malaria J 3:23 4. Jortzik et al. (2011) Biochem J Energy 436:641-50* 5. Allen et al. (2015) FEBS J 282:3808-23*
6. Preuss et al. (2012) J Med Chem 55:7262-72* 7. Berneburg et al. (2022) Antimicrob Agents Chemother (accepted)*
*own puplications
B4Schistosoma mansoni: Biological Targets and Inhibitor Development
Christoph Grevelding
Prof. Dr. Christoph Grevelding
BFS, Institut für Parasitologie
Justus-Liebig-Universität Gießen
Schubertstraße 81
35392 Gießen
Tel.: +49 (0)641-99 38466
Fax: +49 (0)641-99 38469
E-Mail: Christoph.Grevelding(at)vetmed.uni-giessen(dot)de
Project description:
Background: In the human and animal parasite Schistosoma mansoni, the sexual maturation of the female depends on a continuous pairing contact with the male (Abb. 1). Pairing is a prerequisite for egg produc-tion and as such decisive for the pathogenesis of schistosomiasis, which is induced by the eggs. Our research showed that different classes of molecules are involved in organizing reproductive processes in schisto-somes.1 Among others, experiments with kinase inhibitors and by kinase RNA interference demonstrated that kinases regulate cell division, sper-matogenesis, oogenesis, egg production and vitality of schistosomes. For instance, the Abl-tyrosine kinase inhibitor imatinib (cancer drug Glivec) negatively affected reproduction and caused the degradation of the gastrodermis of adult S. mansoni in vitro with lethal consequences.2,3
Besides kinases, which we study in cooperation with the Falcone group4, we investigate further enzymes such as aldehyde dehydrogenases (ALDHs) and an aldehyde reductase (AR). In schistosomes, as in other organisms, these molecules are putatively involved in regulating responses to molecular stress. Inhibiting these molecules might devitalize the parasite. First in vitro experiments with inhibitors against these molecules, like the ALDH inhibitor disulfiram, caused morphologic alterations, a decrease of pairing stability, reduced vitality and finally, the death of adult schistosomes within days in vitro. Helicases might also represent interesting targets as suggested by studies using the RNA helicase-specific inhibitor Silvestrol.5 In adult S. mansoni, Silvestrol reduced the vitality of adult worms in vitro but also stem cell-proliferation in gonad cells (Abb. 2). Together with the working groups of Prof. Schlitzer and Prof. Grünweller (Marburg), we will investigate these potential target molecules and develop synthetic inhibitors6,7 to study their physiological und morphological effects, first in vitro. Further, top candidates will be used for in vivo tests in a rodent infection model.

Abb. 1. Bright-field microscopy of a S. mansoni couple. During the constant pairing contact, the female (arrow) resides within the ventral groove of the male. Pairing is the essential pre-requisite for the production of eggs (stars).
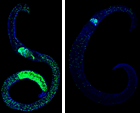
Abb. 2. EdU-staining of female S. mansoni before (left) and after Silvestrol treatment (right). The num-ber of proliferating stem cells (green) is significantly reduced (right).
Scientific goal:
We will clone and characterize two RNA helicases of S. mansoni at the molecular and biochemical levels. In different cooperations within DRUID (see below), we will test the recombinant proteins in enzyme assays against Silvestrol and synthetic derivatives of this substance. Furthermore, we continue our analyses of the recombinantly expressed enzymes SmALDH312 and SmAR, further potential target molecules, and perform enzyme tests with inhibitors. Among these are disulfiram derivatives, which exhibited anti-schistosomal effects in vitro as shown in previous experiments. In addition, we plan to crystallize these molecules for structure analyses. Aim is to develop substances with inhibitor activity against the selected target molecules with high specificity, bioavailability and reduced toxicity for the host.8 Finally, results of the in vitro and in vivo experiments provide a basis for using this knowledge also for other parasitic systems, in which effective compounds will be tested (platform projekt E1).
DRUID Collaboration partners:
A2 AG Grünweller, B3 AG Rahlfs/Kolb/van Zandbergen, B5 AG Schlitzer, E1 Platformprojekt/Häberlein, E3 AG Rahlfs/Przyborski, E5 AG Czermak/Salzig lab
References B4: 1Beckmann et al. (2010) PLoS Pathog 6:e1000769; 2Beckmann et al. (2010) Int J Parasitol 40:521-6; 3Beckmann et al. (2012) Curr Pharm Des 18:3579-94; 4Moreira et al. (2022) Molecules (in press); 5Taroncher-Oldenburg et al. (2021) Microorganisms 9(3):540; 6Peter Ventura et al. (2021) ChemMedChem 14(21):1856-62; 7Peter Ventura et al. (2012) Arch Pharm (Weinheim) 354(12): e2100259; 8Mäder et al. (2018) ChemMedChem 13(22):2374-89.
B5Development of Dithiocarbamates as Anthelminthics and Inhibitors of RNA-helicase eIF4A as potential antiviral Agents
Martin Schlitzer
Prof. Dr. Martin Schlitzer
Institut für Pharmazeutische Chemie
Fachbereich Pharmazie
Philipps-Universität Marburg
Marbacher Weg 6
35037 Marburg
Tel.: +49 (0)6421 28-25840
E-Mail: schlitzer(at)staff.uni-marburg(dot)de
Project description:
Helminthic infections represent a major problem in large parts of the world. In comparison to the burden the arsenal of anthelminthic drugs is rather limited. Using successive cycles of design, synthesis and testing the novel class of dithiocarbamates shall be optimized regarding activity towards different helminths, host toxicity and drug-likeness. Derivatives are synthesized, tested against different helminths and human cell lines. For promising derivatives ADME-parameters are determined and selected compounds could be tested in vivo.
Different viruses pathogenic to humans, as for instance the Ebola- or the SARS-CoV-2 virus, are using host factors for intracellular replication. These host factors therefor represent a target for anti-viral drug design. One of these host factors is the RNA-helicase eIF4A. Based on the crystal structure of the eIF4A-RNA complex appropriate ligands are designed, evaluated by docking and in case of proper fit synthesized. Compounds are tested in regard to their effect on translation efficiency. Active derivatives are subsequently evaluated regarding their effect on virus replication in cell cultures.

Scientific goal:
Both projects should yield compounds which possess the quality of a lead structure or possibly a development candidate
DRUID Collaboration partners:
A2, A3, A4, A7, B2, B7P, C6 NWG, D4, E1, E4, E6
References B5: –
B6 PIdentification of antiviral targets in lipid metabolism
Eva Herker
Prof. Dr. Eva Herker
Institut für Virologie
Philipps-Universität Marburg
Hans-Meerwein-Str. 2
35043 Marburg
Tel.: +49 (0)6421-28-64525
E-Mail: eva.herker(at)uni-marburg(dot)de
Project description:
The human pathogenic flaviviruses Dengue (DENV), Yellow Fever (YFV), Zika (ZIKV), West Nile (WNV) and tick-borne encephalitis virus (TBEV) cause acute infections with severe complications. Fundamental steps in flavivirus replication are closely linked to cellular lipids. These include, among other things, membrane reorganizations for the formation of replication vesicles or capsid envelopment. Flaviviruses alter the lipid composition of the host cell, and the activity of various lipid-metabolizing enzymes is essential for successful replication. For this reason, enzymes from different lipid metabolism pathways represent interesting targets for (pan-) antiflaviviral therapy. However, it has not yet been clarified in detail whether the above-mentioned flaviviruses are dependent on different or similar branches of lipid metabolism.
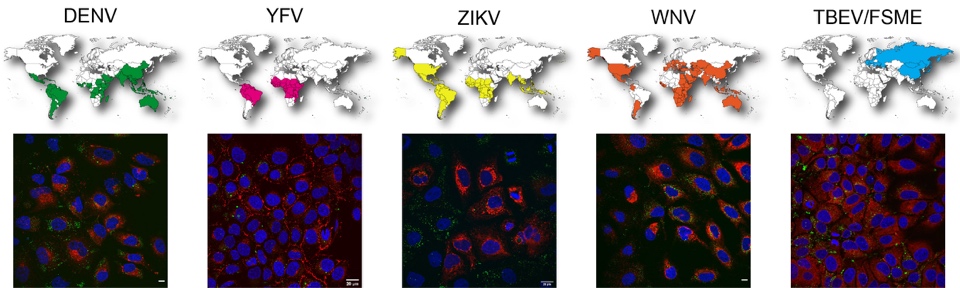
Immunofluorescence microscopy of cells infected with different flaviviruses; red: viral E protein; green: lipid droplets.
Scientific goal:
The project aims to identify antiviral targets in the cellular lipid metabolism. The role of various key enzymes in de novo fatty acid and cholesterol biosynthesis, in phospholipid and neutral lipid metabolism, and of lipid remodeling enzymes in flavivirus replication is analyzed using an shRNA-based screen. In addition, various inhibitors are tested. The results are validated in different cell types and the molecular mechanisms are examined in detail
DRUID Collaboration partners:
B1 Diederich/Kolb, D1 Böttcher-Friebertshäuser/Steinmetzer, E7P Krijnse-Locker, E4 Spengler
References B6P: Herker et al. (2010) Nat Med 16: 1295 2. Harris et al. (2011) J Biol Chem 286: 42615 3. Herker et al. (2012) J Biol Chem 287: 2280 4. Rosch et al. (2016) Cell Rep 16: 3219 5. Hofmann et al. (2018) Biochim Biophys Acta Mol Cell Biol Lipids 1863: 1041 6. Schobel et al. (2018) Sci Rep 8: 3893 7. Lassen et al. (2019) J Cell Sci 132: jcs.217042 8. Bley et al. (2020) Int J Mol Sci 21: 9. Herker et al. (2021) Trends Cell Biol 31: 345 10. Nguyen-Dinh et al. (2021) Cells 10: 2407
B7 PNew ways towards the control of schistosomiasis and echinococcosis
Franco Falcone
Prof. Dr. Franco Falcone
Institut für Parasitologie
BFS - Biomedizinisches Forschungszentrum Seltersberg
Justus-Liebig-Universität Gießen
Schubertstraße 81
35392 Gießen
Tel.: +49 (0)641-99 38030
E-Mail: franco.falcone(at)vetmed.uni-giessen(dot)de
Project description:
Echinococcosis and Schistosomiasis are two co-called Neglected Tropical Diseases, which have a severe impact on the health of affected individuals in endemic countries. Our group is developing new tools that can be used for a better control of these two diseases. For schistosomiasis, we are developing new drugs that target the schistosomal Mitogen Activated Protein Kinases [1], in an effort to find new drugs, which are larvicidal as well as adulticidal. For Echinococcosis, we are developing a novel diagnostic platform based on detection of parasite-specific IgE using humanized IgE reporter cell lines [2-4], which can be used in a variety of formats. We are also working on extending this diagnostic platform to other parasitic infections (e.g. Fasciola, Cysticercosis, Clonorchis, Opisthorchis).
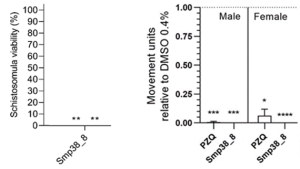
Larvicidal and adulticidal activity of compound 38_8, chosen from a compound library using our bioinformatic approach.
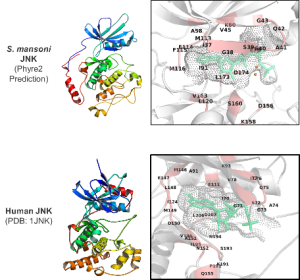
Structural prediction of S. mansoni JNK compared to the human orthologue.
Scientific goal:
The Schistosome project aims to identify lead kinase inhibitor molecules using a bioinformatics in silico analysis pipeline and further develop these into suitable drugs, while in the Echinococcus project we are pursuing a better understanding of the protective immune responses against echinococci and aim to incorporate this knowledge into the development of vaccines and greatly improved, innovative diagnostic technologies.
DRUID Collaboration partners:
A4 Heine lab, B4 Grevelding lab, C6 NWG Häberlein lab, E5 Czermak/Salzig lab
References B7P: [1] *Pereira-Moreira et al. (2020) ACS Omega 5:9064-9070 [2] Kalli M et al. (2020) Sci Rep. 10:18208 [3] Kalli M. et al. (2020) Methods Mol Biol.;2163:155-162. [4] Prakash PS et al. (2021) Parasitol Res.








