Project description:
Background: In the human and animal parasite Schistosoma mansoni, the sexual maturation of the female depends on a continuous pairing contact with the male (Abb. 1). Pairing is a prerequisite for egg produc-tion and as such decisive for the pathogenesis of schistosomiasis, which is induced by the eggs. Our research showed that different classes of molecules are involved in organizing reproductive processes in schisto-somes.1 Among others, experiments with kinase inhibitors and by kinase RNA interference demonstrated that kinases regulate cell division, sper-matogenesis, oogenesis, egg production and vitality of schistosomes. For instance, the Abl-tyrosine kinase inhibitor imatinib (cancer drug Glivec) negatively affected reproduction and caused the degradation of the gastrodermis of adult S. mansoni in vitro with lethal consequences.2,3
Besides kinases, which we study in cooperation with the Falcone group4, we investigate further enzymes such as aldehyde dehydrogenases (ALDHs) and an aldehyde reductase (AR). In schistosomes, as in other organisms, these molecules are putatively involved in regulating responses to molecular stress. Inhibiting these molecules might devitalize the parasite. First in vitro experiments with inhibitors against these molecules, like the ALDH inhibitor disulfiram, caused morphologic alterations, a decrease of pairing stability, reduced vitality and finally, the death of adult schistosomes within days in vitro. Helicases might also represent interesting targets as suggested by studies using the RNA helicase-specific inhibitor Silvestrol.5 In adult S. mansoni, Silvestrol reduced the vitality of adult worms in vitro but also stem cell-proliferation in gonad cells (Abb. 2). Together with the working groups of Prof. Schlitzer and Prof. Grünweller (Marburg), we will investigate these potential target molecules and develop synthetic inhibitors6,7 to study their physiological und morphological effects, first in vitro. Further, top candidates will be used for in vivo tests in a rodent infection model.

Abb. 1. Bright-field microscopy of a S. mansoni couple. During the constant pairing contact, the female (arrow) resides within the ventral groove of the male. Pairing is the essential pre-requisite for the production of eggs (stars).
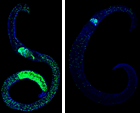
Abb. 2. EdU-staining of female S. mansoni before (left) and after Silvestrol treatment (right). The num-ber of proliferating stem cells (green) is significantly reduced (right).
Scientific goal:
We will clone and characterize two RNA helicases of S. mansoni at the molecular and biochemical levels. In different cooperations within DRUID (see below), we will test the recombinant proteins in enzyme assays against Silvestrol and synthetic derivatives of this substance. Furthermore, we continue our analyses of the recombinantly expressed enzymes SmALDH312 and SmAR, further potential target molecules, and perform enzyme tests with inhibitors. Among these are disulfiram derivatives, which exhibited anti-schistosomal effects in vitro as shown in previous experiments. In addition, we plan to crystallize these molecules for structure analyses. Aim is to develop substances with inhibitor activity against the selected target molecules with high specificity, bioavailability and reduced toxicity for the host.8 Finally, results of the in vitro and in vivo experiments provide a basis for using this knowledge also for other parasitic systems, in which effective compounds will be tested (platform projekt E1).
DRUID Collaboration partners:
A2 AG Grünweller, B3 AG Rahlfs/Kolb/van Zandbergen, B5 AG Schlitzer, E1 Platformprojekt/Häberlein, E3 AG Rahlfs/Przyborski, E5 AG Czermak/Salzig lab
References B4: 1Beckmann et al. (2010) PLoS Pathog 6:e1000769; 2Beckmann et al. (2010) Int J Parasitol 40:521-6; 3Beckmann et al. (2012) Curr Pharm Des 18:3579-94; 4Moreira et al. (2022) Molecules (in press); 5Taroncher-Oldenburg et al. (2021) Microorganisms 9(3):540; 6Peter Ventura et al. (2021) ChemMedChem 14(21):1856-62; 7Peter Ventura et al. (2012) Arch Pharm (Weinheim) 354(12): e2100259; 8Mäder et al. (2018) ChemMedChem 13(22):2374-89.