E3: Protein production, assay development, crystallisation and interaction analysis
Prof. Dr. Jude Przyborski
Interdisziplinäres Forschungszentrum (iFZ)
Justus-Liebig-Universität Gießen
Heinrich-Buff-Ring 26-32
35392 Gießen
Tel.: +49 (0)641-99 39114
E-Mail: jude.przyborski(at)ernaehrung.uni-giessen(dot)de
Dr. Stefan Rahlfs
Biochemie und Molekularbiologie
Justus-Liebig-Universität Gießen
Heinrich-Buff-Ring 26-32
35390 Gießen
Tel.: +49 (0)641-99 39117
E-Mail: stefan.rahlfs(at)ernaehrung.uni-giessen(dot)de
Project description:
Within the framework of this platform project, we offer methods for protein production, for the development of high-throughput assays, for crystallisation and structural analysis as well as for interaction analyses. A variety of optimisation options for cloning/expression and purification of native proteins (affinity chromatography, gel filtration, ion exchange) are available. Once recombinant protein has been obtained, provide support in the development of assay systems up to high throughput formats. Furthermore, the platform proves access to the crystallisation platform at the iFZ. A pipetting and a crystallisation robot as well as starter kits and various additive screens for optimisation are available for crystallisation screens on the nl scale. The system is currently being updated with funding from the latest DRUID funding round. Dr Fritz-Wolf acts as crystallographer and contact person and can carry out the first tests on the crystals. The X-ray sources of the Max Planck Institute for Medical Research (Heidelberg) and the AG Klebe/Heine [A5] (Marburg) are available for this purpose.
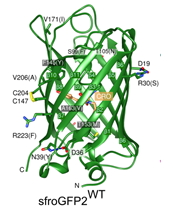
Crystal structure of sfroGFP (Heimsch et al. 2022)
DRUID Collaboration partners:
Protein-Expression: A1, A3, A6NWG, A7, B2, B3, B4, C3
Crystallisation: B2, B3, C3
Assay-Devlopment: A7, B3
References E3: 1. Heimsch et al. (2022) Antioxid & Redox Signal doi: 10.1089/ars.2021.0234 2. Harnischfeger et al. (2020) Electronic Journal of Biotechnology 3. Fritz-Wolf et al. (2011) Nat Commun 2:383 4. Koncarevic et al. (2009) Proc Natl Acad Sci USA 106:13323-8


