D2: HEV, Identification of cellular targets for antiviral strategies-inhibition of virus release by modulation of cholesterol level
Dr. Kai-Henrik Peiffer
Goethe-Universität Frankfurt am Main
Universitätsklinikum
Zentrum der Inneren Medizin
Medizinische Klinik I
Theodor-Stern-Kai 7
60596 Frankfurt am Main
E-Mail: kai-henrik.peiffer(at)kgu(dot)de
Prof. Dr. Eberhard Hildt
Bundesinstitut für Impfstoffe
und biomedizinische Arzneimittel
Paul-Ehrlich-Institut
Paul-Ehrlich-Straße 51-59
63225 Langen
Tel.: +49 (0)6103-77 2140
Fax: +49 (0)6103-77 1234
E-Mail: Eberhard.Hildt(at)pei(dot)de
Prof. Dr. Stefan Zeuzem
Department of Medicine
Goethe University Hospital
Theodor-Stern-Kai 7
60590 Frankfurt am Main
Tel.: +49 (0)69-6301 4544
E-Mail: zeuzem(at)em.uni-frankfurt(dot)de
Project description:
HEV infects more than 20 Mio people per year and concerns industrial nations as well as developing countries. At present there is no specific therapy available.As non-enveloped virus HEV release depends on endodomal processes which represent a promising target for antivirals. In the first funding period the lysosmal degradation of HEV in the endosomal system was identified as a central anchor point for antivirals. In addition to innate immunity cholesterol homeostasis is a a central factor affecting HEV reelase. Based on this the underlying mechanisms are analysed in detalil to identifiy further targets which can be addressed by drug repurposing. Animal models will help to broaden the insight in immunological processes controlling HEV life cycle.
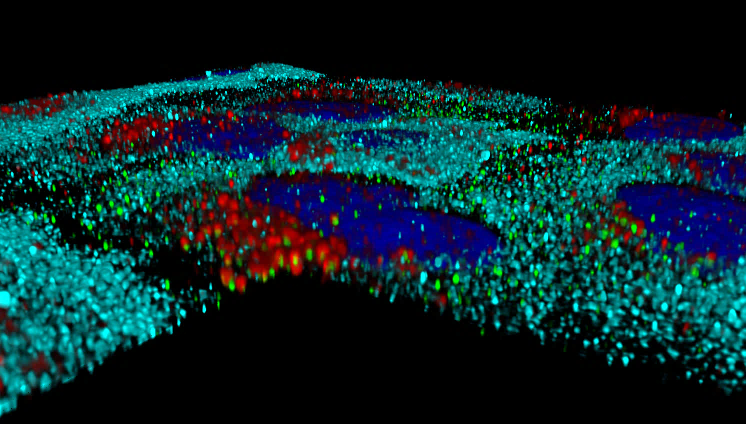
3D reconstruction of subcellular distribution of HEV (green) , GBP-1 (cyan) and lysosomes (red) in interferon -treated cells.
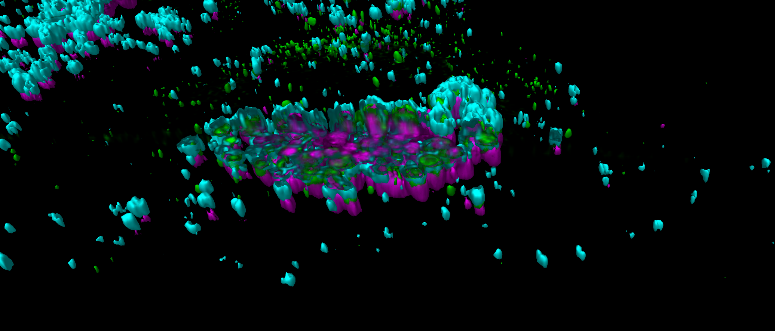
CLSM-based 3D reconstruction of sub-cellular distribution of HEV (green) and lysosomes (cyan) after induction of and cholesterol accumulation (magenta).
Scientific goal:
Inhibtion of the endosomal life cycle of HEV by modulation of cholesterol-dependent regulated target structurs via application of identified bioactive compounds (drug repurposing).
DRUID Collaboration partners:
A2 Grünweller lab, A4 Heine/Reuter lab, B1 Diederich/Kolb lab, B6P Herker lab, C5 Glebe/Geyer lab, D1 Steinmetzer lab
References D2: [1] Glitscher et al. (2018) Viruses 10(6):301; [2] Müller et al. (2020) Antiviral Res 174:104706; [3] Basic et al. (2019) Antiviral Res 172:104644; [4] Glitscher et al. (2021) J Virol doi: 10.1128/JVI.01564-20; [5] Glitscher et al. (2021) Cell Mol Gastroenterol Hepatol, doi:10.1016 /j.jcmgh.2021.02.002; [6] Himmelsbach et al. (2018) Emerg Microbes Infect 7(1):196. [7] Glitscher et al. (2021) Cell Microbiol. 16:e13379



