A1: Protein/protein interactions of Ebola virus proteins as targets for new antiviral strategies
Prof. Dr. Stephan Becker
Institut für Virologie
Philipps-Universität Marburg
Hans-Meerwein-Str. 2
35043 Marburg
Phone: +49 (0)6421-28 66253
E-Mail: becker(at)staff.uni-marburg(dot)de
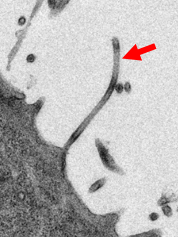
Electronmicroscopic picture of Ebola virus leaving an infected cell (red arrow). ©Schauflinger
Project description
The Ebola virus (EBOV) causes severe fever with extraordinarily high fatality rates. The matrix protein VP40 of EBOV plays key roles for the virus replication cycle and is regulated by homooligomerization. VP40 dimerization is crucial for the protein’s transport towards the plasma membrane where the dimers polymerize resulting in filament formation which enables virus budding. VP40 octamerization results in the down-regulation of viral RNA synthesis. Due to their central role as building blocks of the higher-order oligomers, dimers represent a promising target for antiviral intervention. A fragment-based approach was used to identify hits binding to VP40 crystals which will be developed into lead compounds in order to inhibit VP40 oligomerization.
Scientific goal:
High-resolution crystal Structure of Ebola virus VP40 Dimers. ©Anke Werner
The project aims to develop lead molecules into antiviral compounds using structure-based drug design – a combination of protein crystallography with in silico methods – as well as cell culture experiments under BSL4- conditions for validation.
DRUID collaboration partners:
B1 Diederich/Kolb lab, A4 Heine/Reuter lab, D1 Steinmetzer lab, E3 Rahlfs/Przyborski lab
References A1: 1. *Hartlieb et al. (2007) PNAS 104: 624-9 2. *Hartlieb et al., (2003) J. Biol. Chem. 278: 41830-6 3. *Hoenen et al. (2005) J Virol. 79: 1898-905 4. *Möller et al. 79, 14876-86 (2005) J Virol. 5. Hoenen et al. (2010) J Virol 84: 7053-63. 6. *Gomis-Ruth et al. (2003) Structure 11: 423-33.
* own project-specific preliminary work

